Gene Expression Analysis of Solid Organ Transplants and Validation of Gene Footprints for Transplant Rejection and Tolerance
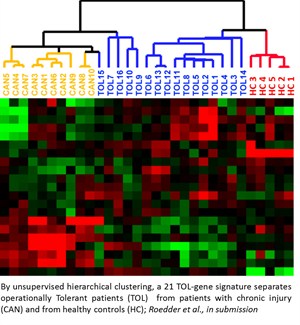 Although recent advances in immunosuppression therapy have enhanced short-term outcome; acute rejection remains an important risk factor for allograft failure. Current management of a transplant patient requires a tissue biopsy as a gold standard for diagnosis, though the process remains limited by sampling error, assessment variability, procedural morbidity and cost. Additionally, renal allograft dysfunction is a relatively insensitive means of detecting early acute rejection such that approximately 10% of patients with clinically normal renal function are found to have evidence of acute rejection on surveillance biopsy. A non-invasive, sensitive and specific means for diagnosing acute rejection, using peripheral blood, urine or any other biofluid, could be used to maintain relatively frequent surveillance of transplant recipients, limiting the need for the invasive biopsy and allowing for proactively immune management to limit graft injury or immunosuppressive morbidity. Transcriptional profiling studies on renal allograft biopsy specimens in our lab have demonstrated substantial, coordinated expression changes in many genes that uniquely identify patients with established acute rejection, as well as other conditions in the differential diagnosis for acute rejection such as chronic rejection, drug toxicity and BK virus nephritis. In general, these changes are felt to be related to the inflammatory infiltrate resident within the kidney and resultant transcriptional changes in renal tissue. However, when these studies have been applied to peripheral blood the diagnostic changes related to acute rejection have been less evident, presumably due to a reduced signal to noise ratio inherent in a site remote from the allograft.In order to increase the sensitivity and specificity of detection for relatively rare biomarkers within heterogeneous samples such as peripheral blood, we have developed a carefully designed and methodological approach that recognizes and accepts the heterogeneity of the sample, its processing and clinical confounders. We have been using gene expression microarrays for unbiased hypothesis generation and QPCR techniques for targeted validation, refine gene markers that can diagnose acute rejection and immune quiescence (operational tolerance) in kidney, heart, and liver transplant patients. These studies have been done in collaboration with KOLs in organ transplantation in the US, EU and Mexico and the kSORT assay is being prospectively validated in a RCT in Europe and being used for an interventional trial in Mexico and in the US (NIH-NIAID-CTOT grant).
Although recent advances in immunosuppression therapy have enhanced short-term outcome; acute rejection remains an important risk factor for allograft failure. Current management of a transplant patient requires a tissue biopsy as a gold standard for diagnosis, though the process remains limited by sampling error, assessment variability, procedural morbidity and cost. Additionally, renal allograft dysfunction is a relatively insensitive means of detecting early acute rejection such that approximately 10% of patients with clinically normal renal function are found to have evidence of acute rejection on surveillance biopsy. A non-invasive, sensitive and specific means for diagnosing acute rejection, using peripheral blood, urine or any other biofluid, could be used to maintain relatively frequent surveillance of transplant recipients, limiting the need for the invasive biopsy and allowing for proactively immune management to limit graft injury or immunosuppressive morbidity. Transcriptional profiling studies on renal allograft biopsy specimens in our lab have demonstrated substantial, coordinated expression changes in many genes that uniquely identify patients with established acute rejection, as well as other conditions in the differential diagnosis for acute rejection such as chronic rejection, drug toxicity and BK virus nephritis. In general, these changes are felt to be related to the inflammatory infiltrate resident within the kidney and resultant transcriptional changes in renal tissue. However, when these studies have been applied to peripheral blood the diagnostic changes related to acute rejection have been less evident, presumably due to a reduced signal to noise ratio inherent in a site remote from the allograft.In order to increase the sensitivity and specificity of detection for relatively rare biomarkers within heterogeneous samples such as peripheral blood, we have developed a carefully designed and methodological approach that recognizes and accepts the heterogeneity of the sample, its processing and clinical confounders. We have been using gene expression microarrays for unbiased hypothesis generation and QPCR techniques for targeted validation, refine gene markers that can diagnose acute rejection and immune quiescence (operational tolerance) in kidney, heart, and liver transplant patients. These studies have been done in collaboration with KOLs in organ transplantation in the US, EU and Mexico and the kSORT assay is being prospectively validated in a RCT in Europe and being used for an interventional trial in Mexico and in the US (NIH-NIAID-CTOT grant).