Clinical & Translational ResearchThe Need for Personalized Medicine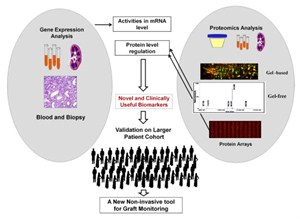 Sensitive and effective biomarkers for different diseases are unmet need. Evolution in high throughput molecular screening methods and developments in bioinformatics tools in recent years have made it possible to conduct comprehensive biomarker discovery efforts in different diseases including organ transplantation. Relentless effort in basic and clinical research have contributed to the identification of potential biomarkers and disease relevant pathways. However, significant work is required before the academic discovery can actually be translated to the patient in the clinic. Integration of de-identified patient demographic data with clinical parameters, our own experimental data and the massive amount of publicly available massive genomic, transcriptomic and proteomic data, allows us to accelerate the power of discovery of new mechanisms and monitoring tools for human diseases. The aim of these studies is to identify potentially important diagnostic, prognostic, and therapeutic markers for disease diagnosis, monitoring and improved therapeutic intervention. Sensitive and effective biomarkers for different diseases are unmet need. Evolution in high throughput molecular screening methods and developments in bioinformatics tools in recent years have made it possible to conduct comprehensive biomarker discovery efforts in different diseases including organ transplantation. Relentless effort in basic and clinical research have contributed to the identification of potential biomarkers and disease relevant pathways. However, significant work is required before the academic discovery can actually be translated to the patient in the clinic. Integration of de-identified patient demographic data with clinical parameters, our own experimental data and the massive amount of publicly available massive genomic, transcriptomic and proteomic data, allows us to accelerate the power of discovery of new mechanisms and monitoring tools for human diseases. The aim of these studies is to identify potentially important diagnostic, prognostic, and therapeutic markers for disease diagnosis, monitoring and improved therapeutic intervention.
Cystinosis Research, A Rare Autosomal Recessive Lysosomal Storage DisorderCystinosis and impaired autophagy/Mitophagy: Cystinosis is a rare autosomal recessive lysosomal storage disorder caused by the mutations in the CTNS gene encoding the lysosomal membrane transporter cystinosin, resulting in the accumulation of toxic metabolite cysteine in the cells. Continuous cystine accumulation eventually leads to multiorgan dysfunction and in the absence of treatment, they usually develop progressive renal failure by the end of the first decade. Treatment with the drug cysteamine depletes the intracellular cystine and if used early in the disease and in high doses it can lower the progression of renal glomerular damage and extra-renal organ injury.
Read more KIDCOV - Assessment of Kidney Injury in Non-Hospitalized in the COVID-19 Pandemic KIDCOV is a study about Kidney Function and the effects of COVID.
The project is led by an outstanding team of clinicians and researchers across five academic medical/research centers from the University of California system, Rush University, and the University of Michigan. The study will enroll and track 2,000 participants, half have tested positive for COVID-19 and the other half have tested negative.
Read More Organ Transplantation ResearchKidney Precision Medicine Project (KPMP)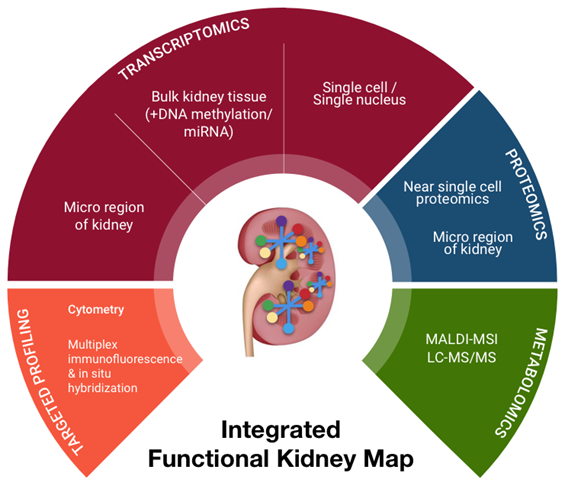 The Kidney Precision Medicine Project (KPMP) is focused on finding new ways to treat acute kidney injury (AKI) and chronic kidney disease (CKD). We will be obtaining kidney tissues from individuals volunteering to participate in KPMP. The tissue will be analyzed in an effort to redefine kidney disease in molecular terms and identify novel targeted therapies. The network will develop state-of-the-art methods to obtain and analyze these biopsies, linking information on cellular programs with kidney structure. We are also developing the next generation of software tools to visualize and understand the various components of these devastating diseases. The Kidney Precision Medicine Project (KPMP) is focused on finding new ways to treat acute kidney injury (AKI) and chronic kidney disease (CKD). We will be obtaining kidney tissues from individuals volunteering to participate in KPMP. The tissue will be analyzed in an effort to redefine kidney disease in molecular terms and identify novel targeted therapies. The network will develop state-of-the-art methods to obtain and analyze these biopsies, linking information on cellular programs with kidney structure. We are also developing the next generation of software tools to visualize and understand the various components of these devastating diseases.
Through collaboration with research teams at UCSF and Stanford, we have developed two multiplex assays (multiplex Immunofluorescence and In Situ Hybridization (mIFISH) and CODEX) that visualize multiple mRNAs and proteins at single cell resolution level. These assays, coupled with advanced high resolution whole slide imaging and sophisticated computer-assisted image analysis, can assess not only the quantitative aspects but also the spatial organizational aspects of the analytes. The in situ assays will be supplemented by additional ancillary tissue-based assays of near single cell proteomics and multiplexed single-cell RNA-Seq (mdrosc RNA-Seq). Gene Expression Analysis of Solid Organ Transplants and Validation of Gene Footprints for Transplant Rejection and Tolerance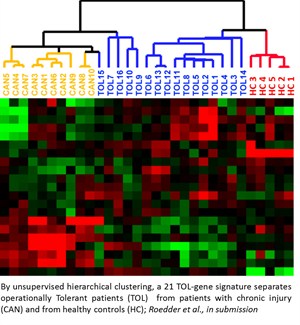 Although recent advances in immunosuppression therapy have enhanced short-term outcome; acute rejection remains an important risk factor for allograft failure. Current management of a transplant patient requires a tissue biopsy as a gold standard for diagnosis, though the process remains limited by sampling error, assessment variability, procedural morbidity and cost. Additionally, renal allograft dysfunction is a relatively insensitive means of detecting early acute rejection such that approximately 10% of patients with clinically normal renal function are found to have evidence of acute rejection on surveillance biopsy. A non-invasive, sensitive and specific means for diagnosing acute rejection, using peripheral blood, urine or any other biofluid, could be used to maintain relatively frequent surveillance of transplant recipients, limiting the need for the invasive biopsy and allowing for proactively immune management to limit graft injury or immunosuppressive morbidity. Transcriptional profiling studies on renal allograft biopsy specimens in our lab have demonstrated substantial, coordinated expression changes in many genes that uniquely identify patients with established acute rejection, as well as other conditions in the differential diagnosis for acute rejection such as chronic rejection, drug toxicity and BK virus nephritis. In general, these changes are felt to be related to the inflammatory infiltrate resident within the kidney and resultant transcriptional changes in renal tissue. However, when these studies have been applied to peripheral blood the diagnostic changes related to acute rejection have been less evident, presumably due to a reduced signal to noise ratio inherent in a site remote from the allograft.In order to increase the sensitivity and specificity of detection for relatively rare biomarkers within heterogeneous samples such as peripheral blood, we have developed a carefully designed and methodological approach that recognizes and accepts the heterogeneity of the sample, its processing and clinical confounders. We have been using gene expression microarrays for unbiased hypothesis generation and QPCR techniques for targeted validation, refine gene markers that can diagnose acute rejection and immune quiescence (operational tolerance) in kidney, heart, and liver transplant patients. These studies have been done in collaboration with KOLs in organ transplantation in the US, EU and Mexico and the kSORT assay is being prospectively validated in a RCT in Europe and being used for an interventional trial in Mexico and in the US (NIH-NIAID-CTOT grant). Although recent advances in immunosuppression therapy have enhanced short-term outcome; acute rejection remains an important risk factor for allograft failure. Current management of a transplant patient requires a tissue biopsy as a gold standard for diagnosis, though the process remains limited by sampling error, assessment variability, procedural morbidity and cost. Additionally, renal allograft dysfunction is a relatively insensitive means of detecting early acute rejection such that approximately 10% of patients with clinically normal renal function are found to have evidence of acute rejection on surveillance biopsy. A non-invasive, sensitive and specific means for diagnosing acute rejection, using peripheral blood, urine or any other biofluid, could be used to maintain relatively frequent surveillance of transplant recipients, limiting the need for the invasive biopsy and allowing for proactively immune management to limit graft injury or immunosuppressive morbidity. Transcriptional profiling studies on renal allograft biopsy specimens in our lab have demonstrated substantial, coordinated expression changes in many genes that uniquely identify patients with established acute rejection, as well as other conditions in the differential diagnosis for acute rejection such as chronic rejection, drug toxicity and BK virus nephritis. In general, these changes are felt to be related to the inflammatory infiltrate resident within the kidney and resultant transcriptional changes in renal tissue. However, when these studies have been applied to peripheral blood the diagnostic changes related to acute rejection have been less evident, presumably due to a reduced signal to noise ratio inherent in a site remote from the allograft.In order to increase the sensitivity and specificity of detection for relatively rare biomarkers within heterogeneous samples such as peripheral blood, we have developed a carefully designed and methodological approach that recognizes and accepts the heterogeneity of the sample, its processing and clinical confounders. We have been using gene expression microarrays for unbiased hypothesis generation and QPCR techniques for targeted validation, refine gene markers that can diagnose acute rejection and immune quiescence (operational tolerance) in kidney, heart, and liver transplant patients. These studies have been done in collaboration with KOLs in organ transplantation in the US, EU and Mexico and the kSORT assay is being prospectively validated in a RCT in Europe and being used for an interventional trial in Mexico and in the US (NIH-NIAID-CTOT grant).
Gene Expression Analysis of Bone Marrow Transplants and Validation of Gene Footprints for Graft vs Host DiseaseGVHD is a clinical problem that has no predictive markers and is a source of high morbidity and mortality for the recipient. Our Lab has used microarrays and QPCR to find a novel gene panel in peripheral blood that can predict the onset of chronic GVHD. These studies are currently being cross-validated in RCT in bone marrow transplants with our collaborators at the Fred Hutchinson Institute and the University of Miami. Proteome and Peptidome Analysis of Serum and Urine of Organ Transplant Patients to Develop Protein Panels for Specific Transplant Injury Phenotypes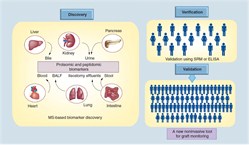
Urine analysis has a unique advantage in kidney transplantation as it reflects graft injury events that occur in the kidney. Non-invasive urinary biomarker identification, specific and sensitive for graft injury diagnosis and prognosis, would eliminate serial protocol biopsies, advance allograft surveillance, and potentially improve graft survival. While gene based biomarker studies in blood samples are underway in the Sarwal Lab, post-translational modifications of the protein product of the gene may be more critical for determining pathogenesis. With recent advances in proteomics methods and analyses, we have undertaken analysis of the ultrafiltrate of the inflamed transplant kidney- the urine- as the biofluid for non-invasive biomarker discovery for different forms of graft injury. We applied shotgun proteomics to map the human urinary proteome to at least 1446 unique urinary proteins. Using a semi quantitative assay with protein-level spectral counts we identified a number of transplant injury specific urinary proteins. This first of its kind study not only identified novel proteins in the urine of renal transplant patients but also help elucidate the increased presence of proteins related with MHC antigens, the complement cascade and extra-cellular matrix. Similar approaches have been applied to the analyses of sera from patients with kidney, heart and lung transplants, and have shown highly specific protein signatures during rejection in each organ. Our Lab has also developed the analysis methodologies for peptidomics of endogenous urinary peptides. This approach provides a tool to examine thousands of small MW endogenous peptides using a top-down approach. This approach identifies peptides in the molecular weight range of 900-5000 Da by tandem mass spectrometry (MS/MS) and can yield the identification of thousands of peptides in a single experiment. These peptides could be used as clinically useful biomarkers for different renal and systemic diseases as they are product of proteolytic degradation. Integrative Approaches to Understanding Chronic Allograft Injury 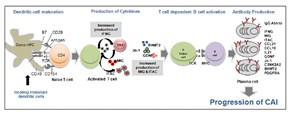 Chronic allograft injury (CAI) is a poorly understood process of accelerated histological injury in the allograft that results in accelerated functional decline and is the major cause of transplanted kidney loss. We have taken an innovative 3 pronged integrated analysis on serial matched biopsy, urine, and serum samples incorporating biopsy microarrays, urinary proteomics and serum protein-arrays collected from the same kidney transplant patients to discover, validate and predict CAI, before irreversible damage occurs, and eventually replace the invasive renal biopsy. Chronic Allograft Injury (CAI) is best considered as a non-specific end-pathway of tubulointerstitial, microvascular and glomerular damage that results from the variety of insults to the transplanted kidney. Chronic allograft injury (CAI) is a poorly understood process of accelerated histological injury in the allograft that results in accelerated functional decline and is the major cause of transplanted kidney loss. We have taken an innovative 3 pronged integrated analysis on serial matched biopsy, urine, and serum samples incorporating biopsy microarrays, urinary proteomics and serum protein-arrays collected from the same kidney transplant patients to discover, validate and predict CAI, before irreversible damage occurs, and eventually replace the invasive renal biopsy. Chronic Allograft Injury (CAI) is best considered as a non-specific end-pathway of tubulointerstitial, microvascular and glomerular damage that results from the variety of insults to the transplanted kidney.
The resultant fibrotic lesion is a manifestation of the allografts limited healing repertoire in response to this varying injury, and its evolution remains the main clinical barrier to sustained graft survival, despite improvements in immunosuppression and decreased acute graft loss. The use of protocol biopsies in long-term monitoring studies has found that allograft damage is common across all renal transplants. This damage is time-dependent, progressive and underestimated by the measurement (rise) of serum creatinine alone. CAI has many immune and non-immune causes that have been implicated, which include acute rejection, subclinical rejection, calcineurin inhibitor nephrotoxicity, ischemia/reperfusion injury and pre-existing donor disease.
The corresponding molecular pathways leading to allograft injury are less well defined and are limited to a few studies. These cross-sectional (clinically indicated samples) or surveillance biopsy studies have demonstrated fibrogenic gene expression, although when derived from end-stage grafts, the information becomes less clinically useful in a chronic setting. None have focused on the early post-transplant period. There have also not been any analysis of corresponding biopsy, urine and blood samples by different approaches, in the same patient, which can significantly enhance the discovery process for critically regulated pathways in CAI.
Our research has focused to fill these gaps in the understanding of CAI, by using an integrative comprehensive analysis of the allograft and recipient transcriptome (biopsy, urine and serum) matched with clinical data on protocol biopsies; beginning early post-transplant right through to 2 years post-transplant. We will use this new understanding to develop clinically relevant non-invasive biomarkers of CAI that can enhance patient monitoring for customized management and graft surveillance. Integrated Bioinformatics in Organ Transplantation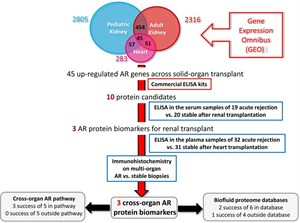 We are conducting a large-scale genome-wide screening study across injury datasets in organ transplantation, using highly annotated genomic and proteomic datasets in the Sarwal Lab as well as within the public domain. With our expertise in translational bioinformatics to integrate genomic, proteomic, metabolic data from multiple sources, we are developing comprehensive strategy for integrated bioinformatics analysis of archived renal transplant genomic and proteomic datasets available in the Sarwal Lab together with publically available data pertaining to different types of organ transplants (kidney, heart, lung, liver, pancreas, and intestine) and different bacterial, viral and fungal infections. These studies will lead to improved understanding of the cross- talk between innate, allo and heterologous immunity in organ transplantation. We are conducting a large-scale genome-wide screening study across injury datasets in organ transplantation, using highly annotated genomic and proteomic datasets in the Sarwal Lab as well as within the public domain. With our expertise in translational bioinformatics to integrate genomic, proteomic, metabolic data from multiple sources, we are developing comprehensive strategy for integrated bioinformatics analysis of archived renal transplant genomic and proteomic datasets available in the Sarwal Lab together with publically available data pertaining to different types of organ transplants (kidney, heart, lung, liver, pancreas, and intestine) and different bacterial, viral and fungal infections. These studies will lead to improved understanding of the cross- talk between innate, allo and heterologous immunity in organ transplantation.
|